Hey there! This is Xin Cao, a Master of Philosophy in Computational Biology at The Chinese University of Hong Kong, Shenzhen (CUHKSZ) I also aquired my Bachelor of Science in Data Science & Big Data Technology degree at CUHKSZ.
I am currently under the supervision of Prof. Hsien-Da HUANG in the Warshel Institute for Computational Biology, focusing on Transcriptional Factor (TF) Activity and Regulatory Network Prediction based on Bayesian & Deep Learning Models, and miRNA Data. I have also worked with Prof. Gang CAO at the Shenzhen University of Advanced Technology, where I led a project on Protein Protein Interaction Prediction (PPI) using Graph Neural Networks (GNNs). In the past, I have also followed Prof. Zhizheng Wu of CUHKSZ in topics on Music Tracks Generation via Latent Diffusion Models (LDM) and Audio Enhancement. My first research experience was supervised by Prof. Feng ZHENG at SUSTech and contributed some works on Multimodal Computer Vision (CV) Adversarial Robustness, and did surveys on AI music.
Growing up in Cold Spring Harbor (CSHL), my interests and instincts for Biological Sciences have been cultivated greatly; and I chose DS as my major because I foresaw what impact Big Data could have in future lives: it would be the “20th-century-CS-language in the 21st century”. Combining my major with my interests, I am highly dedicated and motivated in exploring frontier fields and potential breakthroughs in Biological Sciences, and have been at the edge of my seat to see what sparks DS/AI can ignite in BioScience!
As the 2024 Nobel Prizes came out and Hold Thorp (Editor-in-Chief of Science) praised AlphaFold as the greatest achievement in AI, I have become even more convinced and confident that the research fields I’ve chosen are the correct ones:
- AI-4-BioScience / Biomedical AI: Artificial Intelligence can and should be utilized to explore Biological Science Data Patterns and contribute to the well-being of humans.
- Neuromorphic Computing (Brain-Like ANNs): Artificial Neural Networks (ANNs) have been blazing through these recent years, but named after human brains, they don’t actually simulate the workflow of human Neural Networks at all! After all, the result after eras of natural selection is what’s truly meant to be intelligent.
- AI Music (Non-generative): Tracks Seperation, Audio Enhancement, Arrangements Assisting.
I am actively seeking interesting research opportunities and Ph.D. positions in the future. If you are looking for a dedicated and self-motivated candidate, please DO contact me at xincao@link.cuhk.edu.cn !
For the final part (though I’d definitely prefer putting this paragraph in the front) I would love to talk about the major achievements aside of my academic pursuits. I am the core player / captain of the CUHKSZ / Harmonia College Soccer Team ⚽️, 2022 champion 🏆 of Shenzhen City, and I’ve also formed multiple bands and we wrote many original songs (covering genres like Hardcore Punk, Blues Rock, R&B, Jazz-Hiphop, Contemporary Folk, Post Rock, etc.) as a guitarist 🎸 keyboard 🎹 and lead singer 🎤; Led Zeppelin and Guns’n’Roses are among my favorite bands. Additionally, I attained ABRSM Grade 5 in Piano & Music Theory at Los Angeles by the age of 10, and achieved First Place in the 2021 CUHKSZ Singing Competition (Duet). Though extracurricular, I firmly believe these experiences may enhance my leadership & teamwork skills professionally, while also fostering creativity and open-mindedness in research.
🔥 News
- 2025.09 🎉 1-st Author Paper ACCEPTED by Briefings in Bioinformatics!!!
- 2025.06: 🧪 Was assigned a new project: Enzyme Function Prediction via Deep Learning Models.
- 2025.03: 📄 Submitted my 1st paper as 1st Author, Enhancing Cross Domain Protein and Peptide Interaction with Re-trained Deep_Learning Models, to Briefing in Bioinformatics, currently Under Review.
- 2024.10: 💬 Participated in the 6th Warshel Institue Meeting of Computational Biology and Bioinformatics, engaged and consulted with various esteemed academics.
- 2024.10: 📈 Joined Prof. Hsien-Da HUANG’s Team: began work on TF Activity Prediction with miRNA and Bayesian Models.
- 2024.09: 🎉 4th Place in the ICSR 2024 Robot Design Competition: presented PASIR, a multimodal emtion prediction robot using BCI-driven emotion models.
- 2024.08: 🎤 Delivered a Speech at the 2024 Opening Ceremony as the Student Representative of Harmonia College, awarded a scholarship of ¥10,000 RMB.
- 2024.05: 🧬 Started research on PPI Prediction via GNNs with Prof. Prof. Gang CAO at Shenzhen University of Advanced Technology.
- 2024.04: 🏅 Ranked Global Top 10 in the 2024 Bremen Big Data Challenge, led the project under supervision of Prof. Haizhou LI
- 2023.10: 🏆 Champion of 2022 Shenzhen Soccer League, Runner-up of 2023 League, Core Player of school team.
- 2024.11: 🎸 Upcoming performance at HOULIVE, Shenzhen’s No.1 livehouse.
📜 Working Papers and Academic Presentations
[1] X. Cao, J. Li, F. Meng, Y. Zou, Z. Wan, K. Xiao (2024). Deep Learning-Based Prediction of Protein-Protein Interactions on Short Protein Datasets. (Manuscript in preparation)
[2] Y. Chen, X. Cao, Y. Pan, X. Chen, A. Gao, P. Chao, S. Huang, M. Li, H. Huang (2024). Bayesian Approach Towards Transcriptional Factor Activity Prediction based on miRNA Data. (Model in Development)
[3] Z. Wan, A. Gao, P. Chao, X. Cao, J. Song, M. Ran (2024). Variation Method for Map Matching. (Manuscript in preparation)
[4] X. Cao, J. Li (2024). PASIR: A Proactive Social Interaction Robot Empowered by Multimodal Data Fusion and BCI-Driven Emotion Prediction Models. International Conference on Social Robotics (ICSR). (Oral presentation)
[5] Reviewed 1 paper for SIGKDD Conference (2025) on behalf of official program committee member: Large Language Models for Next Group Point-of-Interest Recommendation.
📝 Research Experience
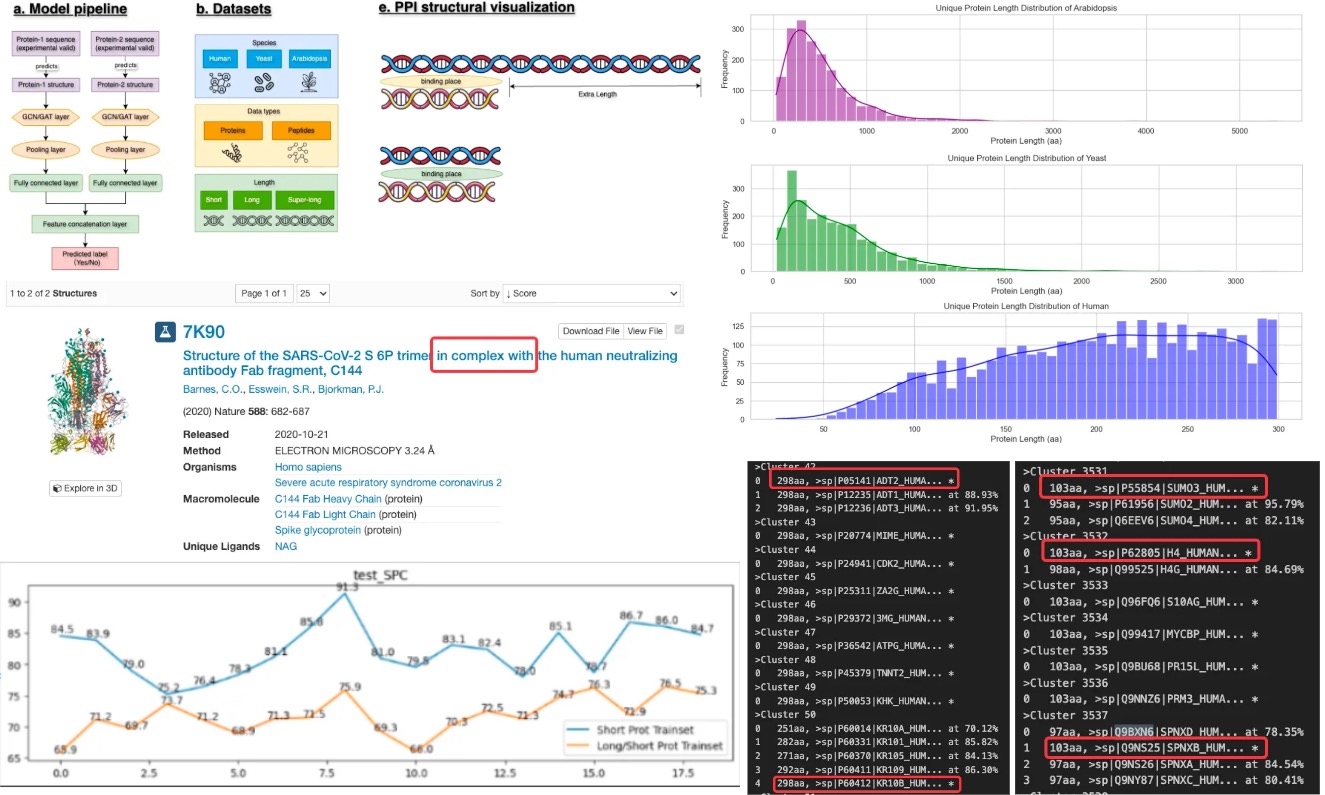
Protein Interaction Prediction: Short Protein Dataset Investigation and Peptide Analysis
Shenzhen University of Advanced Technology, Supervised by Prof. Gang Cao
- Summary: Investigated protein-protein interaction (PPI) prediction model architecture and datasets, proposing the use of short protein sequences to optimize feature learning & computational efficiency. Built a cross-species, non-redundant protein and peptide database and visualized PPI features.
- Methods and Tools: Utilized PyTorch, Tensorflow for model construction (GIN, GAT, VQ-VAE, etc.) and finetuning; CDHIT, MatPlotLib, Linux awk for data processing; AlphaFold2, seqvec, LSTM for protein structure information construction.
- Key Outcomes: Found out training on short protein datasets could improve novel PPI prediction, and finetuning on long+short datasets could improve model performance. Achieved 99.94% accuracy and 98.85% F1 score, establishing a high-performance benchmark for future research.
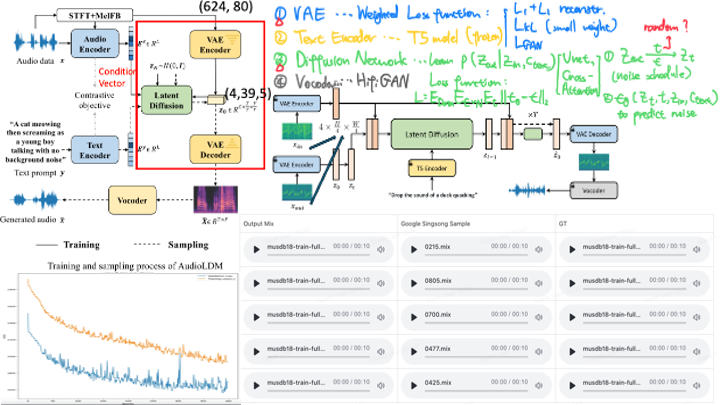
Drum Track Generation & Audio Enhancement
The Chinese University of Hong Kong, Shenzhen, Supervised by Prof. Zhizheng Wu
- Data Collection and Analysis: Collected and analyzed model codes and datasets for audio enhancement and generation tasks. Compared different models’ effectiveness on track generation tasks, concluding that Latent Diffusion and Transformer models performed best. Trained and generated over 90 drumbeat segments for pure music tracks using the Latent Diffusion model.
- Data Processing and Augmentation: Utilized Demucs software to seperate drum tracks from complete music pieces. Adjusted the ratio of different musical styles in dataset to generate more diverse and optimal drumbeats.
- Model Deployment: Responsible for the local deployment and testing of the Demucs model (hybrid transformer architecture) as part of the research team’s ongoing investigation into audio editing tasks.

Multimodal CV Adversarial Robustness and AI Music Survey
SUSTech, Supervised by Prof. Feng ZHENG
- Evaluation of System Robustness: Investigated and organized various metrics to assess and enhance the robustness of image-text retrieval systems. Participated in data preprocessing, model debugging, and evaluating model performance, focusing on the impact of preprocessing techniques on evaluation results.
- Research and Model Re-implementation: Independently researched the fields of Optical Music Recognition (OMR) and AI music generation. Re-implemented and adjusted codes from recent papers, optimizing label data to improve model accuracy. Delivered technical analyses and summaries during team meetings.
🎖 Honors and Awards
- 2024.09 : 4th Place, ICSR 2024 Robot Design Competition
- 2024.08 : Outstanding Leader (¥10,000 RMB), 2023 Chinese University of Hong Kong, Shenzhen Harmonia Annual Scholarship
- 2024.04 : Global Top10 Ranking, 2024 Bremen Big Data Challenge
- 2024.03 : Silver, 7th Aunual Sports Festival of Chinese University of Hong Kong, Shenzhen (Team Captain)
- 2023.01 : Champion, 2022 Shenzhen Annual Soccer League (Team Core Player)
🎓 Educations
- 2020.09 - 2024.12 (now) : The Chinese Uiversity of Hong Kong, Shenzhen; Shenzhen China
- B.Sc., Data Science & Big Data Technology, School of Data Science
- Related Courses : Advanced Machine Learning, Reinforcement Learning, Deep Learning and Applications, Speech and Natural Language Processing, Data Mining, Database Systems, Bioinformatics, Bayesian Statistics, Stochastic Simulation, Stochastic Processes, Optimization, Operations Management, C/C++
- 2008.09 - 2012.12 : Doyle & Southgrove; CA, United States & NY, United States
- Honors: Project Beyond Class (school Top 10% students)
🎤 Invited Talks
- 2024.09 : 16th International Conference on Social Robotics (ICSR 2024)
- 2021.03 : Harmonia College Opening Ceremony, Student Representative of 2024
💻 Proffesional Experiences
- 2021.06 - 2021.09 : Algorithm Engineer, Kunyu Biotechnology Co., Ltd., Wuhan, China.
- Gene Alignment Program Development: Developed gene alignment program using Python and BLAST in two rounds to identify uniquely high-repeating DNA fragments in telomere and centromere regions of target chromosone. Quantitatively analyzed and optimized the threshold changes on final results, obtained high quality probes and an Invention Patent.
- Program Optimization: Assessed and reformed algorithm logic, comparatively experimented different toolkits. Reduced execution time from 48h to under 10h, memory usage by threefold.
- Data Processing and Report Writing: Processed and analyzed output data in Linux, integrated biological information to select detectable sequences without fold spatial structures. Compiled final sequences and wrote user-friendly manual.
🚀 Additional Skills
- 🛠️ Technical : Python, Linux, PyTorch, Tensorflow, SQL, Spark, RegEx, LaTeX, C/C++, Java, Stable Diffusion, Ollama
- 📖 English : Five years of school in America (Native Speaker); TOEFL 100 (Listening: 29)
- 🎵 Music : Experienced in Logic Pro, Reaper, Lumafusion; Lead vocal, lead guitar, keyboard, captain of original band; 1st place in University Singing Competition (Duet); University choir Tenor; ABRSM Grade 5 in piano
- ⚽️ Soccer : Captain of college team; Core player of university team; Champion of Shenzhen University soccer League
